About the DANIO-CODE programme
First release of the DANIO-CODE track hub with a thousand epigenomics tracks is open! #IZCF2018, #ZencodeITN, #zebrafish
Check it out https://danio-code.zfin.org ! Find your favourite gene, its expression, its true promoter or predict its enhancers and noncoding RNAs around it!
https://danio-code.zfin.org/
DANIO-CODE is an international collaborative effort that aims to annotate the functional elements of the zebrafish genome.
Zebrafish are increasingly used to successfully model human disease, to screen drugs and to study environmental toxicity in addition to being a model for embryonic development. Zebrafish genome is the third most complete but further annotation is essential to better realize the power of this model organism in diverse areas of biomedical research and to further strengthen its role in biomedical research.
DANIO-CODE partners work together to provide a central resource of publicly available data and genome annotation of the zebrafish genome. DANIO-CODE combines a large variety of genome wide datasets including protein-coding and non-coding transcribed genome elements, non-coding functional elements such as cis-regulatory modules and associated epigenetic features.
DANIO-CODE was established at a workshop held in Imperial College London in December 2014 (see meeting report published in Zebrafish) where members of the zebrafish genomics research community and previous contributors to ENCODE and FANTOM genome annotation projects identified key aims.
The DANIO-CODE key aims
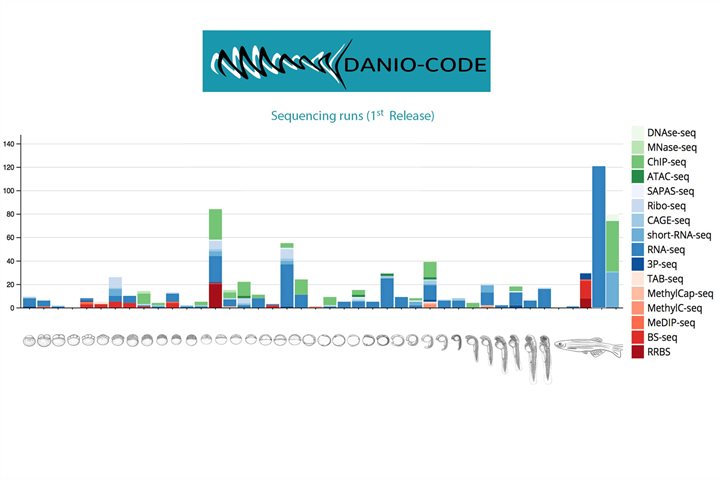
- Establishment of a Data Coordination Centre for standardised data input into a shared Data Resource. This will provide centralised and convenient access and visualisation of currently inaccessible zebrafish genomics datasets such as ChIP-seq, CAGE-seq, RNA-seq, ATAC-seq, Ribo-seq, meDIP-seq. As a first step, published data is being reanalyzed for GRCz10 by standard pipelines with the aim that data will become mineable through a Track hub hosted by ZFIN and managed by DANIO-CODE partners.
- Identification and generation of common policies on data standardization, integration and data sharing.
- Seeking and obtaining funding through joint ventures for genomics resource generation and management identified as community priorities
- Among the future aims is to establish a consortium agreement and a platform for sharing unpublished data and to work together in community inspired annotation efforts.