Replisome disassembly: mechanism and importance for cell biology (funded by Wellcome Trust)
To ensure faultless duplication of the whole genome, DNA replication initiates from thousands of origins of replication. An origin fires when the replicative helicases are activated and start to unwind double stranded DNA creating two DNA replication forks. The progressing replication forks move through the chromatin until they encounter forks from the neighbouring origins. When the forks converge the termination of replication forks happen: the helicases pass each other, synthesis of DNA is completed, the replisomes disassemble and topisomerase II resolves the daughter DNA molecules (see model below).
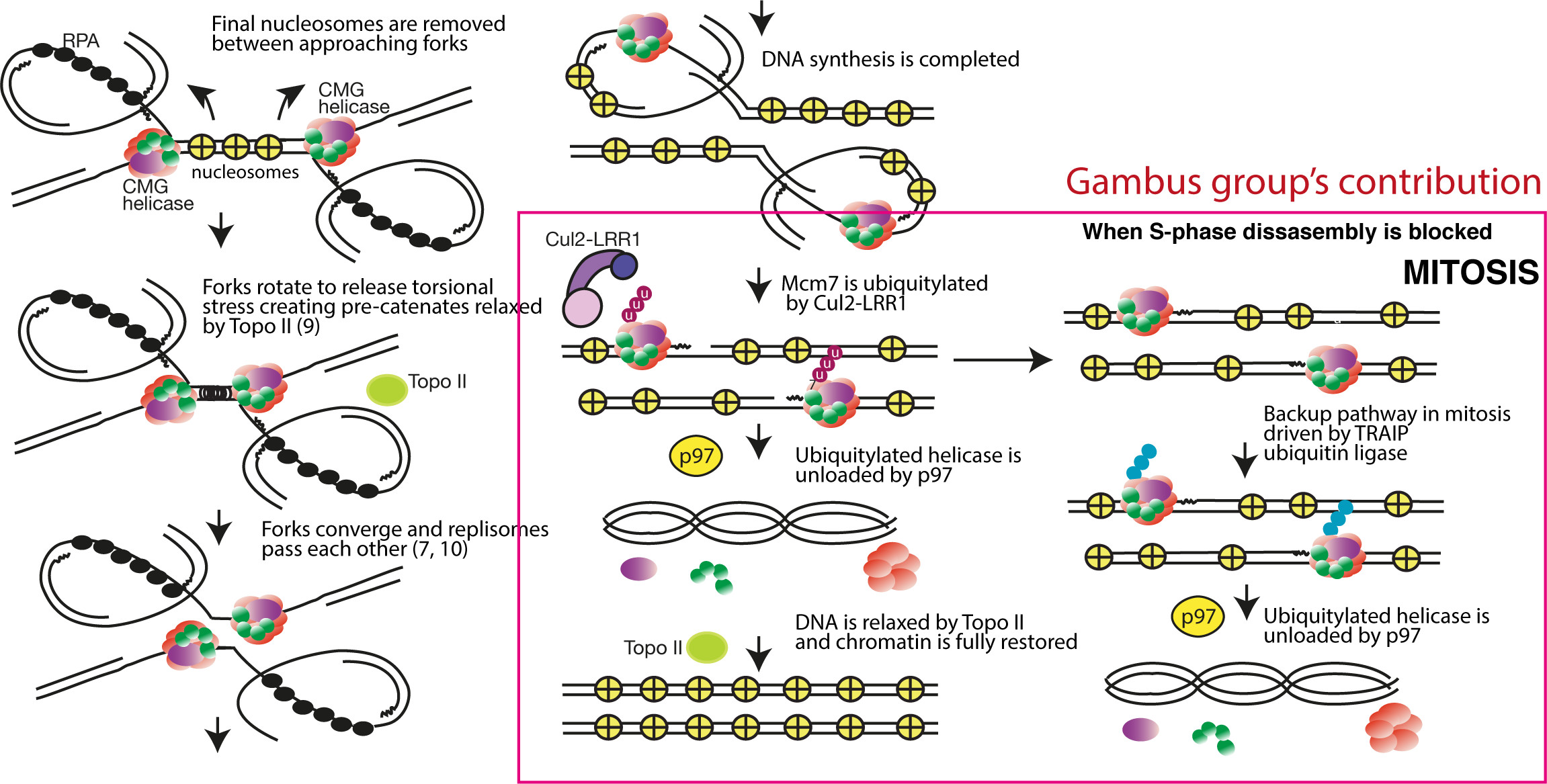
Figure: Model of termination of eukaryotic DNA replication forks based on work conducted by groups of Johannes Walter, Karim Labib and Aga Gambus.
We have identified the first elements of the mechanism of replisome disassembly at the termination of eukaryotic DNA replication and the role of ubiquitylation in this process. We found that only one subunit of the replicative helicase, Mcm7, is polyubiquitylated at termination and that following this ubiquitylation, the helicase is removed from chromatin in a p97/VCP segregase-dependent manner. As the helicase forms the organising centre of the replisome, its removal leads to disassembly of the remaining replication machinery (Moreno et al. Science 2014). Subsequently, we identified the Cul2Lrr1 ubiquitin ligase as the enzyme ubiquitylating Mcm7 in higher eukaryotes (Sonneville et al. NCB 2017). Finally, any replisomes that are not unloaded in S-phase are unloaded in mitosis in a back-up pathway driven by activity of TRAIP ubiquitin ligase (Priego Moreno et al. Life Sci Alliance 2019).
We believe that disruption of the replisome disassembly process is detrimental to human health and that understanding the basis of this process will provide valuable targets that could be therapeutically exploited. We have, however, barely begun to understand the mechanism behind this process. Our efforts now focus, therefore, on determining the detailed mechanism and regulation of this process both in Xenopus laevis egg extract and in human cells. We aim also to understand what are the consequences of disruption of this process.
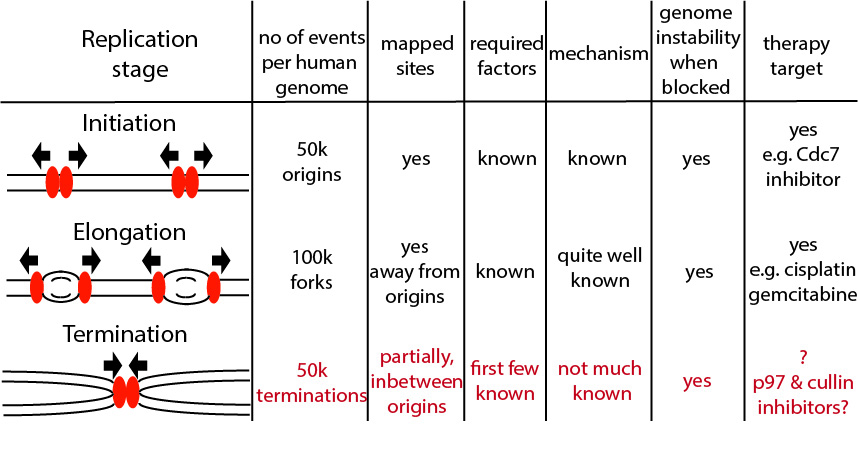
Figure: The importance of studying termination of DNA replication. Over the 65 years of DNA replication research we understand quite a lot about initiation and elongation stages of DNA replication, but the termination stage remained understudied until last five years.
Activation and regulation of TRAIP ubiquitin ligase (funded by BBSRC)
We have shown that TRAIP ubiquitin ligase interacts with post-termination replisome in S-phase but it is efficiently ubiquitylating Mcm7 within the replisome only in mitosis (Priego Moreno et al. Life Sci Alliance 2019). Interestingly, Mcm7 in ubiquitylated with K63 or K6 chains in mitosis and these types of chains can lead to mitotic replisome disassembly.
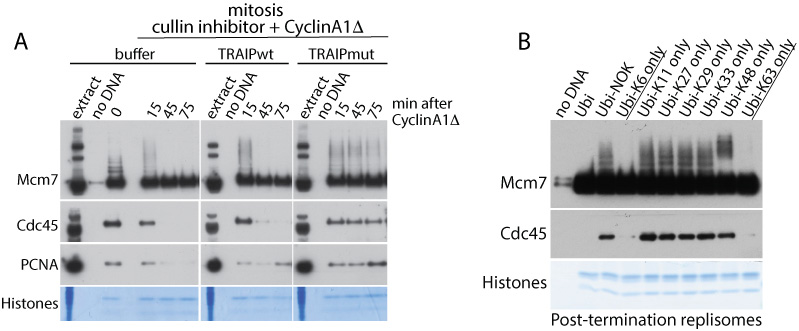
Figure: (A) The activity of TRAIP ubiquitin ligase is essential for unloading of the replisome in mitosis as enzymatic dead mutant of TRAIP (TRAIPmut) blocks unloading of replisome (represented by Cdc45 subunit). (B) Ubiquitylated Mcm7 and Cdc45 are unloaded only when K6 or K63 chains can form on Mcm7 in mitosis.
Our aim is to understand how TRAIP is activated and regulated to productively ubiquitylate Mcm7 only in mitosis and only when replication fork approaches DNA damage during S-phase.
The role of SUMO in regulation of chromosomal replication
The process of sumoylation resembles closely this of ubiquitylation, but the role of SUMO modification during DNA replication and damage are much less understood. We aim therefore to identify replication factors modified my SUMO during the process of replication and characterise the function of these modifications.
Single-Molecule Termination (SinMolTermination, funded by Marie Sklodowska Curie Fellowship for Dr Neville Gilhooly)
SinMolTermination will develop a real-time and super resolution single-molecule imaging platform (Figure) to monitor the progression and termination of replication forks and directly observe protein dynamics within the replisome.
Using this platform we aim to:
- Determine the fate of CMG during termination.
- Define the spatial and temporal conditions upon which Mcm7 becomes ubiquitylated.
Discover how misregulation of termination can give rise to replication stress.
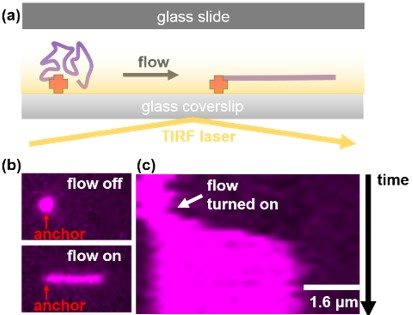
Figure: Combining microfluidics and TIRF microscopy to visualise and manipulate single DNA molecules. (a) DNA molecules that are anchored inside a microfluidic flow cell can be extended using hydrodynamic flow and imaged using TIRFM. (b) Micrograph of a fluorescently stained DNA molecule where flow is turned on and off. Notice the molecule elongates under flow. (c) Kymograph of the DNA molecule shown in (b) showing that the molecule rapidly elongates when flow is turned on.