Genome Structure and Regulation
Our research into gene regulation and epigenetics interfaces with cancer cell and stem cell biology, viral oncology (EBV and HPV), development and genome biology. We use both functional genomics and high throughput methods to determine how genetic abnormalities unravel the complex regulatory networks that underlie cancer and normal development. In parallel we are studying the gene regulatory mechanisms that control the normal development and differentiation of stem cells.
Current research funding includes a £1.3 million grant from the MRC to the Bonifer and Cockerill laboratory to test combinations of drugs that target the signalling molecule FLT3 together with the transcription factor RUNX1 in Acute Myeloid Leukaemia, as well as other project grants from the MRC, BHF, BBSRC and EU Horizon programmes.
Lead

Professor Ferenc Mueller
Professor in Developmental Genetics
View profile
Research groups
Spotlight on marrow failure in a zebrafish model of GATA2 deficiency
The Monteiro group employed a zebrafish fish model to replicate what happens in patients who inherit the loss of one allele of the GATA2 gene who develop bone marrow failure and leukaemia. Zebrafish lacking a transcriptional enhancer needed for efficient GATA2 expression exhibit a gradual loss of myeloid blood cells, a skewing towards red blood cells, and an AML-like syndrome.
Cell Reports 42:112571 (2023). Christopher B Mahony, Lucy Copper, Pavle Vrljicak, Boris Noyvert, Chrystala Constantinidou, Sofia Browne, Yi Pan, Claire Palles, Sascha Ott, Martin R Higgs, Rui Monteiro. Lineage skewing and genome instability underlie marrow failure in a zebrafish model of GATA2 deficiency. doi: https://doi.org/10.1016/j.celrep.2023.112571

GATA2 deficiency is a rare genetic bone marrow disorder in which patients are at increased risk of blood cancers. This is likely due to (i) GATA2-deficient blood stem cells in bone marrow producing significantly fewer immune cells used to fight infection, and (ii) the GATA2 gene mutation resulting in impaired activity of genes that are involved in repairing ongoing damage to their DNA.
In humans, GATA2 haploinsufficiency due to mutations in coding or enhancer regions causes hematopoietic disorders collectively referred to as GATA2 deficiency syndromes. 75% of patients with inherited germline GATA2 mutations develop early-onset myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML).
In the zebrafish model employed here, both alleles of an enhancer controlling the gata2a locus were deleted. This generated a bona fide model of GATA2 deficiency that shows marrow hypocellularity, neutropenia, increased susceptibility to infections, and development of an AML-like phenotype in the adult kidney marrow.
Spotlight on global analysis of enhancers that control blood cell development
New study uncovers the information encoded in our DNA required for the transcriptional control of blood cell development from embryonic stem cells
This publication from the Bonifer laboratory describes a novel genome-wide screen that identified and functionally characterised distinct subsets of gene regulatory elements that direct the different stages of the development of blood cells from embryonic stem cells (ESC)
Nature Communications 14:267 (2023). Edginton-White B, A Maytum, SG Kellaway, DK Goode, P Keane,. . . C Bonifer. A genome-wide relay of signalling-responsive enhancers drives hematopoietic specification.
doi: 10.1038/s41467-023-35910-9

This study of the genome investigated the functions of all of the regulatory elements within our DNA that are required to express our genes in the right cells, at the right levels, and at the right time. These regulatory elements are called enhancers and are scattered over large distances within the the 3 billion bases of DNA that make up the genome. Enhancers also function to integrate multiple highly specific intrinsic and extrinsic signals, whereby most regulatory elements only function in a subset of cells. The Bonifer laboratory developed a whole genome assay able to define the precise time course of activation of enhancers, and their responses to external signals, during the stepwise development of blood cells from ESCs. These analyses identified thousands of differentially active enhancers able to stimulate a promoter across different stages of blood cell development from ESCs. It also showed that blood cell-specific gene expression is controlled by the concerted action of thousands of differentiation stage-specific sets of enhancers and promoters, many of which respond to the cytokine signals promoting cell differentiation towards blood cells. This work highlights the mechanisms of how and where extrinsic signals program a cell type-specific regulatory landscape driving hematopoietic differentiation.
This methodology, which can now be adapted to any ESC-derived cell type, also provides a way to investigate how gene expression is sometimes perturbed by mutations in enhancers or external signals which cause disease. To understand how genes respond to outside signals and are deregulated in disease, we need to know where these enhancer elements are, when they are active and how they function.
Spotlight on Cancer Genomics
Nature Genetics 51:151-162 (2019). Assi SA, MR Imperato, DJL Coleman, A Pickin, S Potluri, A Ptasinska, . . . PN Cockerill, C Bonifer. Subtype-specific regulatory network rewiring in acute myeloid leukemia.
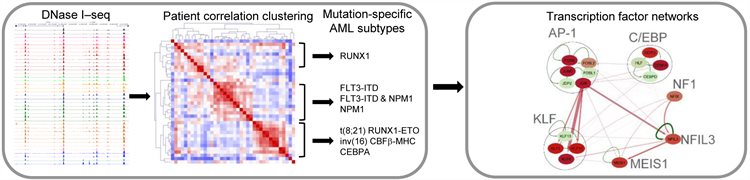
A study by Constanze Bonifer and Peter Cockerill has revealed the roles that different types of gene mutations play in causing acute myeloid leukaemia. Epigenetic profiling of regulatory elements revealed that mutation-specific subsets of AML have distinct patterns of gene expression. Each subset was controlled by distinct gene regulatory networks linked to mutations in transcription factors and signalling molecules This research brings us one step closer to being able to provide tailored and targeted treatment specific to individual patients, increasing their chances of survival.
Selected Highlights from Gene Structure and Regulation
Cell Reports 42:112571 (2023). Christopher B Mahony, Lucy Copper, Pavle Vrljicak, Boris Noyvert, Chrystala Constantinidou, Sofia Browne, Yi Pan, Claire Palles, Sascha Ott, Martin R Higgs, Rui Monteiro. Lineage skewing and genome instability underlie marrow failure in a zebrafish model of GATA2 deficiency. doi: https://doi.org/10.1016/j.celrep.2023.112571
Blood 2022019138 (2023). Clarke ML, RB Lemma, DS Walton, G Volpe, B Noyvert, OS Gabrielsen and J Frampton. MYB insufficiency disrupts proteostasis in hematopoietic stem cells leading to age-related neoplasia. doi: 10.1182/blood.2022019138
Nature Commun 14:267 (2023). Edginton-White B, A Maytum, SG Kellaway, DK Goode, P Keane,. . . C Bonifer. A genome-wide relay of signalling-responsive enhancers drives hematopoietic specification.
Dev Cell 58:155-170 e158 (2023). Hadzhiev Y, L Wheatley, L Cooper, F Ansaloni, C Whalley, Z Chen, . . . A Beggs, F Muller. The miR-430 locus with extreme promoter density forms a transcription body during the minor wave of zygotic genome activation.
Blood doi: 10.1182/blood.2021015036 (2022). Tirtakusuma R, K Szoltysek, P Milne, V Grinev, A Ptasinska, PS Chin, . . . C Bonifer, O Heidenreich, S Bomken. Epigenetic regulator genes direct lineage switching in MLL/AF4 leukaemia.
Elife 11 (2022). Vilaplana-Lopera N, V Cuminetti, R Almaghrabi, G Papatzikas, AK Rout, M Jeeves, . . . P Garcia. Crosstalk between AML and stromal cells triggers acetate secretion through the metabolic rewiring of stromal cells.
Nat Genet 54:1037-1050 (2022). Baranasic D, M Hortenhuber, PJ Balwierz, T Zehnder, AK Mukarram, C Nepal, . . . F Muller. Multiomic atlas with functional stratification and developmental dynamics of zebrafish cis-regulatory elements.
Cancer Lett 501:172-186 (2021). Gleneadie HJ, AH Baker, N Batis, J Bryant, Y Jiang, SJH Clokie, H Mehanna, P Garcia, DMA Gendoo. . . JL Parish, FL Khanim, M Wiench. The anti-tumour activity of DNA methylation inhibitor 5-aza-2'-deoxycytidine is enhanced by the common analgesic paracetamol through induction of oxidative stress.
Front Immunol 12:642807 (2021). Bevington SL, R Fiancette, DW Gajdasik, P Keane, JK Soley, CM Willis, . . . PN Cockerill. Stable Epigenetic Programming of Effector and Central Memory CD4 T Cells Occurs Within 7 Days of Antigen Exposure In Vivo.
Life Science Alliance 4 (2021). Kellaway SG, P Keane, B Edginton-White, K Regha, E Kennett and C Bonifer. Different mutant RUNX1 oncoproteins program alternate haematopoietic differentiation trajectories.
Cell Reports 34:108759 (2021). Wang J, P Rojas, J Mao, M Muste Sadurni, O Garnier, S Xiao, . . . MR Higgs, P Garcia, M Saponaro. Persistence of RNA transcription during DNA replication delays duplication of transcription start sites until G2/M.
Frontiers in Immunology 12:642807 (2021). Bevington SL, R Fiancette, DW Gajdasik, P Keane, JK Soley, CM Willis, . . . PN Cockerill. Stable Epigenetic Programming of Effector and Central Memory CD4 T Cells Occurs Within 7 Days of Antigen Exposure In Vivo.
EMBO Reports 22:e51120 (2021). Blakemore D, N Vilaplana-Lopera, R Almaghrabi, E Gonzalez, M Moya, C Ward, . .A Gambus, E Petermann, GS Stewart, P Garcia. MYBL2 and ATM suppress replication stress in pluripotent stem cells.
Scientific Reports 11:3009 (2021). Halsall JA, S Andrews, F Krueger, CE Rutledge, G Ficz, W Reik and BM Turner. Histone modifications form a cell-type-specific chromosomal bar code that persists through the cell cycle.
Cell Reports 35:109010 (2021). Potluri S, SA Assi, PS Chin, DJL Coleman, A Pickin, S Moriya, . . . PN Cockerill, C Bonifer. Isoform-specific and signaling-dependent propagation of acute myeloid leukemia by Wilms tumor 1.
J General Virology 101:873-883 (2020). Hollingworth R, GS Stewart and RJ Grand. Productive herpesvirus lytic replication in primary effusion lymphoma cells requires S-phase entry.
EMBO J:e105220 (2020). Bevington SL, P Keane, JK Soley, S Tauch, DW Gajdasik, R Fiancette, . . . PN Cockerill. IL-2/IL-7-inducible factors pioneer the path to T cell differentiation in advance of lineage-defining factors. doi: 10.15252/embj.2020105220
Cell Rep 31:107748 (2020). Bevington SL, STH Ng, GJ Britton, P Keane, DC Wraith and PN Cockerill. Chromatin Priming Renders T Cell Tolerance-Associated Genes Sensitive to Activation below the Signaling Threshold for Immune Response Genes.
Commun Biol 3:71 (2020). Dobrzycki T, CB Mahony, M Krecsmarik, C Koyunlar, R Rispoli, J Peulen-Zink, . . . R Monteiro. Deletion of a conserved Gata2 enhancer impairs haemogenic endothelium programming and adult Zebrafish haematopoiesis.
Haematologica (2020). Kellaway SG, P Keane, E Kennett and C Bonifer. RUNX1-EVI1 disrupts lineage determination and the cell cycle by interfering with RUNX1 and EVI1 driven gene regulatory networks. DOI: 10.3324/haematol.2019.241885
Life Sci Alliance 3 (2020). Kwon SY, K Massey, MA Watson, T Hussain, G Volpe, CD Buckley, . . . P Badenhorst. Oxidised metabolites of the omega-6 fatty acid linoleic acid activate dFOXO.
Cell Rep 31:107691 (2020). Nafria M, P Keane, ES Ng, EG Stanley, AG Elefanty and C Bonifer. Expression of RUNX1-ETO Rapidly Alters the Chromatin Landscape and Growth of Early Human Myeloid Precursor Cells.
Nucleic Acids Res 48:8374-8392 (2020). Wragg JW, L Roos, D Vucenovic, N Cvetesic, B Lenhard and F Muller. Embryonic tissue differentiation is characterized by transitions in cell cycle dynamic-associated core promoter regulation.
J Virol 10.1128 (2020). Molloy DP and RJ Grand. Structural determinants within the adenovirus early region 1A protein spacer region necessary for tumourigenesis.
Nat Genet 51:151-162 (2019). Assi SA, MR Imperato, DJL Coleman, A Pickin, S Potluri, A Ptasinska, . . . PN Cockerill, C Bonifer. Subtype-specific regulatory network rewiring in acute myeloid leukemia.
Nat Commun 10:3577 (2019). Bonkhofer F, R Rispoli, P Pinheiro, M Krecsmarik, J Schneider-Swales, IHC Tsang, . . . R Monteiro, T Peterkin, R Patient. Blood stem cell-forming haemogenic endothelium in zebrafish derives from arterial endothelium.
Nat Commun 10:691 (2019). Hadzhiev Y, HK Qureshi, L Wheatley, L Cooper, A Jasiulewicz, H Van Nguyen, . . . F Muller. A cell cycle-coordinated Polymerase II transcription compartment encompasses gene expression before global genome activation.
Cell Rep 28:3022-3031 e3027 (2019). Ptasinska A, A Pickin, SA Assi, PS Chin, L Ames, R Avellino, . . . C Bonifer. RUNX1-ETO Depletion in t(8;21) AML Leads to C/EBPalpha- and AP-1-Mediated Alterations in Enhancer-Promoter Interaction.
Cancer Cell 34:674-689 e678 (2018). de Boer B, J Prick, MG Pruis, P Keane, MR Imperato, J Jaques, . . . C Bonifer, JJ Schuringa. Prospective Isolation and Characterization of Genetically and Functionally Distinct AML Subclones.
Cancer Cell 34:626-642 e628 (2018). Martinez-Soria N, L McKenzie, J Draper, A Ptasinska, H Issa, S Potluri, . . . C Bonifer, O Heidenreich. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation.
Cell Rep 24:1496-1511 e1498 (2018). Ward C, G Volpe, P Cauchy, A Ptasinska, R Almaghrabi, D Blakemore, . . . P Garcia. Fine-Tuning Mybl2 Is Required for Proper Mesenchymal-to-Epithelial Transition during Somatic Reprogramming.
J Immunol 199:2652-2667 (2017). Brignall R, P Cauchy, SL Bevington, B Gorman, AO Pisco, J Bagnall, . . . P Cockerill, P Paszek. Integration of Kinase and Calcium Signaling at the Level of Chromatin Underlies Inducible Gene Activation in T Cells.
J Virol 91 (2017). Harris L, L McFarlane-Majeed, K Campos-Leon, S Roberts and JL Parish. The Cellular DNA Helicase ChlR1 Regulates Chromatin and Nuclear Matrix Attachment of the Human Papillomavirus 16 E2 Protein and High-Copy-Number Viral Genome Establishment.
Cell Rep 19:1654-1668 (2017). Loke J, SA Assi, MR Imperato, A Ptasinska, P Cauchy, Y Grabovska, . . . C Bonifer. RUNX1-ETO and RUNX1-EVI1 Differentially Reprogram the Chromatin Landscape in t(8;21) and t(3;21) AML.
Nucleic Acids Res 45:9874-9888 (2017). Stanulovic VS, P Cauchy, SA Assi and M Hoogenkamp. LMO2 is required for TAL1 DNA binding activity and initiation of definitive haematopoiesis at the haemangioblast stage.
Sci Rep 7:11148 (2017). Volpe G, DS Walton, DE Grainger, C Ward, P Cauchy, D Blakemore, . . . J Frampton. Prognostic significance of high GFI1 expression in AML of normal karyotype and its association with a FLT3-ITD signature.
EMBO J 35:515-535 (2016). Bevington SL, P Cauchy, J Piper, E Bertrand, N Lalli, RC Jarvis, . . . PN Cockerill. Inducible chromatin priming is associated with the establishment of immunological memory in T cells.
PLoS Genet 12:e1005969 (2016). Kwon SY, V Grisan, B Jang, J Herbert and P Badenhorst. Genome-Wide Mapping Targets of the Metazoan Chromatin Remodeling Factor NURF Reveals Nucleosome Remodeling at Enhancers, Core Promoters and Gene Insulators.
Epigenetics Chromatin 9:52 (2016). Wiersma M, M Bussiere, JA Halsall, N Turan, R Slany, BM Turner and KP Nightingale. Protein kinase Msk1 physically and functionally interacts with the KMT2A/MLL1 methyltransferase complex and contributes to the regulation of multiple target genes.
Leukemia 10:1038 (2016). Clarke M, G Volpe, L Sheriff, D Walton, C Ward, W Wei, . . . J Frampton. Transcriptional regulation of SPROUTY2 by MYB influences myeloid cell proliferation and stem cell properties by enhancing responsiveness to IL-3.
Dev Cell 36:572-587 (2016). Goode DK, N Obier, MS Vijayabaskar, ALM Lie, AJ Lilly, R Hannah, . . . C Bonifer. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation.
Dev Cell 38:358-370 (2016). Monteiro R, P Pinheiro, N Joseph, T Peterkin, J Koth, E Repapi, . . . R Patient. Transforming Growth Factor beta Drives Hemogenic Endothelium Programming and the Transition to Hematopoietic Stem Cells.
Cell Rep 12:821-836 (2015). Cauchy P, SR James, J Zacarias-Cabeza, A Ptasinska, MR Imperato, SA Assi, . . . PN Cockerill. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature.
J Virol 89:4770-4785 (2015). Paris C, I Pentland, I Groves, DC Roberts, SJ Powis, N Coleman, . . . JL Parish. CCCTC-Binding Factor Recruitment to the Early Region of the Human Papillomavirus 18 Genome Regulates Viral Oncogene Expression.
Nat Commun 6:7203 (2015). Regha K, SA Assi, O Tsoulaki, J Gilmour, G Lacaud and C Bonifer. Developmental-stage-dependent transcriptional response to leukaemic oncogene expression.
Nature 507:381-385 (2014). Haberle V, N Li, Y Hadzhiev, C Plessy, C Previti, C Nepal, . . . F Muller, B Lenhard. Two independent transcription initiation codes overlap on vertebrate core promoters.
Cell reports 8:1974-1988 (2014). Ptasinska A, SA Assi, N Martinez-Soria, MR Imperato, J Piper, P Cauchy, . . . C Bonifer. Identification of a Dynamic Core Transcriptional Network in t(8;21) AML that Regulates Differentiation Block and Self-Renewal.
Development 141:715-724 (2014). Roberts JA, I Miguel-Escalada, KJ Slovik, KT Walsh, Y Hadzhiev, R Sanges, . . . F Muller. Targeted transgene integration overcomes variability of position effects in zebrafish.
Leukemia 27:661-670 (2013). Clarke M, S Dumon, C Ward, R Jager, S Freeman, B Dawood, . . . J Frampton, P Garcia. MYBL2 haploinsufficiency increases susceptibility to age-related haematopoietic neoplasia.
Genome Res 23:1938-1950 (2013). Nepal C, Y Hadzhiev, C Previti, V Haberle, N Li, H Takahashi, . . . F Muller. Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis.
Leukemia 27:1487-1496 (2013). Volpe G, DS Walton, W Del Pozzo, P Garcia, E Dasse, LP O'Neill, . . . J Frampton, S Dumon. C/EBPalpha and MYB regulate FLT3 expression in AML.