DNA Replication, Stability and Repair
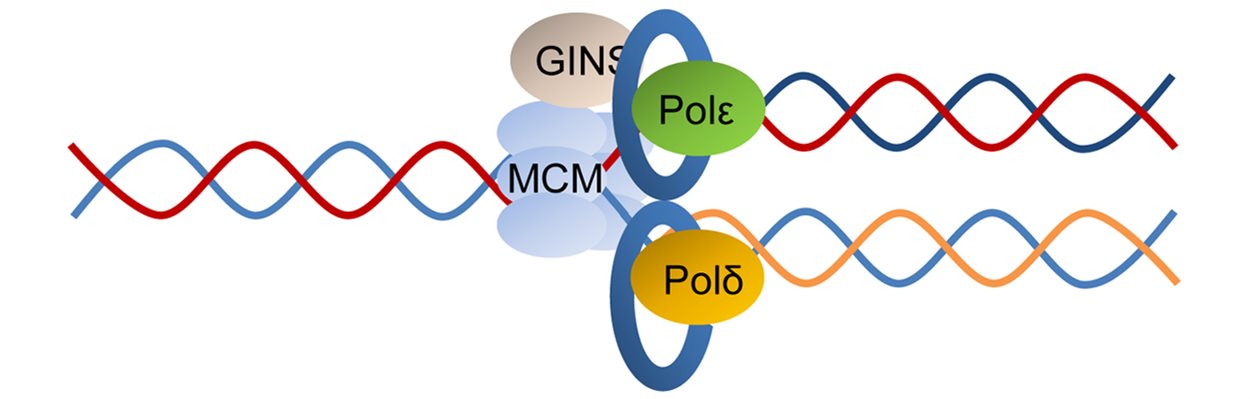
Our research is defining the normal mechanisms of DNA replication and repair, with the aim of understanding how it is deregulated in cancer cells. Maintaining the integrity of our DNA is central to preventing and treating cancer. Mutations in genes that control cell growth can initiate cancer whereas mutations in genes that repair DNA can lead to further mutations in other genes that promote cancer. This is why mutations in DNA repair genes often underlie earlier-onset cancers. Researchers within ICGS are investigating the impact of specific classes of gene mutations, such as ATM, BRCA1, and MYBL2 on genome integrity. We also examine the impact that processes such as transcription and replication have on genome stability.
Over the last decade this theme has been highly successful at attracting major programme and project grant funding. In 2022 Jo Morris secured more than £4.9 million from the Wellcome Trust, the MRC and Horizon Europe to better understand DNA replication and repair. Eva Petermann and Jo Parish were also awarded £1.2 million by the MRC to better understand the role of oncogene activation and genomic instability in cancer initiation.
Theme Lead Theme Deputy Lead

Professor Jo Morris
Professor of Molecular Genetics
View profile

Professor Jason Parsons
Professor of Radiobiology
View profile
Research groups
Spotlight on the role of DONSON in kick-starting DNA replication at origins of replication
Molecular Cell Oct 10:S1097-2765(23)00761-X (2023). Cvetkovic MA, P Passaretti, A Butryn, A Reynolds-Winczura, G Kingsley, A Skagia, . . .G Stewart, A Gambus and A Costa. The structural mechanism of dimeric DONSON in replicative helicase activation.
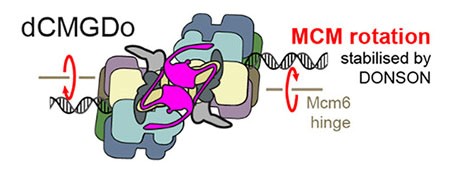
A collaboration between Aga Gambus in Birmingham and Alessandro Costa in London, with contributions from Grant Stewart, Birmingham, has revealed new insights into how DNA replication helicases switch from an inactive form to an active form at DNA origins of replication. High resolution electron microscopy showed how dimers of the protein DONSON (shown here in purple in the centre) bind in between two inactive DNA replication helicases at DNA origins and load symmetrically two helicase activators: the GINS complex. This leads to a conformational change in the complexes, resulting in rotation of the MCM helicases, and the initiation of DNA replication.
Spotlight on Drugs that promote spinal cord repair after injury
Science Advances 8 (2022) eabq2611.Taylor MJ, Thompson AM, Alhajlah S, Tuxworth RI, Ahmed Z. Inhibition of Chk2 promotes neuroprotection, axon regeneration, and functional recovery after CNS injury.
Clinical and Translational Medicine 12 (2022) e962. Ahmed Z, Tuxworth RI. The brain-penetrant ATM inhibitor, AZD1390, promotes axon regeneration and functional recovery in preclinical models of spinal cord injury.
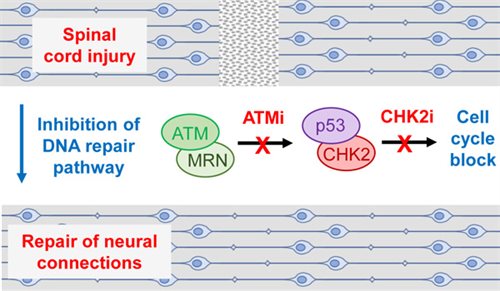
Ground-breaking recent studies from the Ahmed and Tuxworth laboratories have shown that 2 drugs designed to cure cancer can now potentially be used to promote spinal cord and optic nerve repair after injury. Anti-cancer drugs that target either ATM or CHK2 in the DNA repair pathway promote the survival of neurons and the re-growth of axons across sites of nerve damage. DNA double strand breaks in damaged neurons normally trigger a DNA damage response that blocks regrowth of axons and often results in cell death. Remarkably, inhibition of the ATM/CHK2 pathway led to a return of normal transmission of nerve impulses across damaged spinal cords or optic nerves in models of injury in animals where electrical transmission across damaged neurons had been blocked. When these drugs were given within 24 hours of spinal cord injury in animal models, they promoted full recovery of hind limb function and sensation within a few weeks. These drugs are already in development or in use for cancer and may in the future be investigated for their potential use to promote recovery from spinal cord injury in people.
Spotlight on protein modification crosstalk during DNA repair
Nature Communications 12: 6313 (2021). M-P Sanchez-Bailon M-P, S-Y Choi, ER Dufficy, K Sharma, GS McNee, E Gunnel, K Chiang, D Sahay, S Maslen, GS Stewart, JM Skehel, I Dreveny and CC Davies. Arginine methylation and ubiquitylation crosstalk controls DNA end-resection and homologous recombination repair.
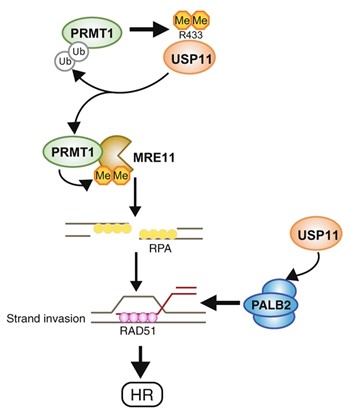
PRMT1 is an arginine methyltransferase that promotes genome stability and is upregulated in many cancers. Here, the group of Clare Davies identify the deubiquitylating enzyme USP11 as a new PRMT1 substrate, and that methylation of USP11 is required for double strand break repair. They also show that PRMT1 is a substrate for USP11, and that deubiquitylation of PRMT1 promotes PRMT1 activity towards MRE11, a component of the MRN complex, leading to efficient DNA end resection at break points. This study demonstrates the complexity of post-translational modifications that safe-guards our genome against DNA damage, but also suggests that hyper-activation could lead to chemo-resistance in cancer cells following chemo-therapy.
Spotlight on BRCA1 and DNA Replication
Nature 571: 521-527 (2019). Daza-Martin M, Starowicz K, Jamshad M, Tye S, Ronson GE, MacKay HL, Chauhan AS, Walker AK, Stone HR, Beesley JFJ, Coles JL, Garvin AJ, Stewart GS, McCorvie TJ, Zhang X, Densham RM, JR Morris. Isomerization of BRCA1-BARD1 promotes replication fork protection.
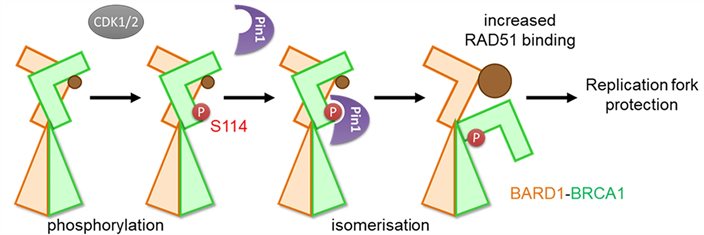
BRCA1 is a gene required for genome stability and is a frequent target of mutations in breast cancer. Here the group of Jo Morris shows that the functional activation of the BRCA1 protein involves a conformational change in its structure.
BRCA1 phosphorylation by CDK1/2 followed by isomerization by PIN1 enhances the ability of BARD1 to associate with RAD51 (brown) and thereby promotes replication fork protection.
Selected Highlights from DNA Replication, Stability and Repair
J Clin Invest 132 (2022). Abu-Libdeh B, SS Jhujh, S Dhar, JA Sommers, A Datta, GM Longo, . . . GS Stewart. RECON syndrome is a genome instability disorder caused by mutations in the DNA helicase RECQL1.
Mol Cell 82:1924-1939 e1910 (2022). Bayley R, V Borel, RJ Moss, E Sweatman, P Ruis, A Ormrod, . . . MR Higgs. H3K4 methylation by SETD1A/BOD1L facilitates RIF1-dependent NHEJ.
J Cell Sci 135 (2022). Faulkner EL, JA Pike, RM Densham, E Garlick, SG Thomas, RK Neely and JR Morris. Imaging nanoscale nuclear structures with expansion microscopy.
J Biol Chem 298:102234 (2022). Tarcan Z, D Poovathumkadavil, A Skagia and A Gambus. The p97 segregase cofactor Ubxn7 facilitates replisome disassembly during S-phase.
Viruses 13 (2021). Abualfaraj T, NC Hagkarim, R Hollingworth, L Grange, S Jhujh, GS Stewart and RJ Grand. The Promotion of Genomic Instability in Human Fibroblasts by Adenovirus 12 Early Region 1B 55K Protein in the Absence of Viral Infection.
EMBO Rep 22:e51120 (2021). Blakemore D, N Vilaplana-Lopera, R Almaghrabi, E Gonzalez, M Moya, C Ward, . . .A Gambus, E Petermann, GS Stewart, P Garcia. MYBL2 and ATM suppress replication stress in pluripotent stem cells.
Nat Comm 12: 6313 (2021). M-P Sanchez-Bailon M-P, S-Y Choi, ER Dufficy, K Sharma, GS McNee, … CC Davies. Arginine methylation and ubiquitylation crosstalk controls DNA end-resection and homologous recombination repair.
Nat Commun 11:5863 (2020). Piberger AL, A Bowry, RDW Kelly, AK Walker, D Gonzalez-Acosta, LJ Bailey, . . . E Petermann. PrimPol-dependent single-stranded gap formation mediates homologous recombination at bulky DNA adducts.
J Gen Virol 101:873-883 (2020). Hollingworth R, GS Stewart and RJ Grand. Productive herpesvirus lytic replication in primary effusion lymphoma cells requires S-phase entry.
Nature 571:521-527 (2019). Daza-Martin M, K Starowicz, M Jamshad, S Tye, GE Ronson, HL MacKay, . . . JR Morris. Isomerization of BRCA1-BARD1 promotes replication fork protection.
Genes Dev 33:333-347 (2019). Garvin AJ, AK Walker, RM Densham, AS Chauhan, HR Stone, HL Mackay, . . . JR Morris. The deSUMOylase SENP2 coordinates homologous recombination and nonhomologous end joining by independent mechanisms.
Cancer Res 78:5767-5779 (2018). Bayley R, D Blakemore, L Cancian, S Dumon, G Volpe, C Ward, . . . MR Higgs, GS Stewart, E Petermann, P Garcia. MYBL2 supports DNA double strand break repair in haematopoietic stem cells.
Cell Rep 25:2061-2069 e2064 (2018). Bowry A, AL Piberger, P Rojas, M Saponaro and E Petermann. BET Inhibition Induces HEXIM1- and RAD51-Dependent Conflicts between Transcription and Replication.
Mol Cell 71:25-41 e26 (2018). Higgs MR, K Sato, JJ Reynolds, S Begum, R Bayley, A Goula, . . . GS Stewart. Histone Methylation by SETD1A Protects Nascent DNA through the Nucleosome Chaperone Activity of FANCD2.
Nat Commun 9:746 (2018). Ronson GE, AL Piberger, MR Higgs, AL Olsen, GS Stewart, PJ McHugh, . . . E Petermann, ND Lakin. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation.
Nat Commun 9:229 (2018). Uckelmann M, RM Densham, R Baas, HHK Winterwerp, A Fish, TK Sixma and JR Morris. USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A.
Nature 559:285-289 (2018). Zimmermann M, O Murina, MAM Reijns, A Agathanggelou, R Challis, Z Tarnauskaite, . . . T Stankovic, AP Jackson, D Durocher. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions.
Mol Cell 65:900-916 e907 (2017). Clarke TL, MP Sanchez-Bailon, K Chiang, JJ Reynolds, J Herrero-Ruiz, TM Bandeiras, . . . CC Davies. PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination.
J Virol 91 (2017). Campos-Leon K, K Wijendra, A Siddiqa, I Pentland, KM Feeney, A Knapman, . . . JL Parish. Association of Human Papillomavirus 16 E2 with Rad50-Interacting Protein 1 Enhances Viral DNA Replication.
Cell Rep 21:3498-3513 (2017). Chiang K, AE Zielinska, AM Shaaban, MP Sanchez-Bailon, J Jarrold, TL Clarke, . . . CC Davies. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression.
Nat Genet 49:537-549 (2017). Reynolds JJ, LS Bicknell, P Carroll, MR Higgs, R Shaheen, JE Murray, . . . GS Stewart. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism.
Nat Cell Biol 19:468-479 (2017). Sonneville R, SP Moreno, A Knebel, C Johnson, CJ Hastie, A Gartner, . . . K Labib. CUL-2LRR-1 and UBXN-3 drive replisome disassembly during DNA replication termination and mitosis.
Cell 168:843-855 e813 (2017). Williamson L, M Saponaro, S Boeing, P East, R Mitter, T Kantidakis, . . . JQ Svejstrup. UV Irradiation Induces a Non-coding RNA that Functionally Opposes the Protein Encoded by the Same Gene.
Nat Commun 7:13087 (2016). Kotsantis P, LM Silva, S Irmscher, RM Jones, L Folkes, N Gromak and E Petermann. Increased global transcription activity as a mechanism of replication stress in cancer.
PLoS Genet 12:e1005945 (2016). Byrd PJ, GS Stewart, A Smith, C Eaton, AJ Taylor, C Guy, . . . AM Taylor. A Hypomorphic PALB2 Allele Gives Rise to an Unusual Form of FA-N Associated with Lymphoid Tumour Development.
Nat Struct Mol Biol 23:647-655 (2016). Densham RM, AJ Garvin, HR Stone, J Strachan, RA Baldock, M Daza-Martin, . . . JR Morris. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection.
Nat Genet 48:36-43 (2016). Harley ME, O Murina, A Leitch, MR Higgs, LS Bicknell, G Yigit, . . . AP Jackson. TRAIP promotes DNA damage response during genome replication and is mutated in primordial dwarfism.
Genes Dev 30:408-420 (2016). Kantidakis T, M Saponaro, R Mitter, S Horswell, A Kranz, S Boeing, . . . JQ Svejstrup. Mutation of cancer driver MLL2 results in transcription stress and genome instability.
Blood 127:582-595 (2016). Kwok M, N Davies, A Agathanggelou, E Smith, C Oldreive, E Petermann, . . . T Stankovic. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells.
Dev Cell 38:358-370 (2016). Monteiro R, P Pinheiro, N Joseph, T Peterkin, J Koth, E Repapi, . . . R Patient. Transforming Growth Factor beta Drives Hemogenic Endothelium Programming and the Transition to Hematopoietic Stem Cells.
Mol Cell 59:462-477 (2015). Higgs MR, JJ Reynolds, A Winczura, AN Blackford, V Borel, ES Miller, . . . GS Stewart. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks.
Science 346:477-481 (2014). Moreno SP, R Bailey, N Campion, S Herron and A Gambus. Polyubiquitylation drives replisome disassembly at the termination of DNA replication.
Cell 157:1037-1049 (2014). Saponaro M, T Kantidakis, R Mitter, GP Kelly, M Heron, H Williams, . . . JQ Svejstrup. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress.
EMBO J 32:1556-1567 (2013). Davies CC, A Chakraborty, ME Diefenbacher, M Skehel and A Behrens. Arginine methylation of the c-Jun coactivator RACO-1 is required for c-Jun/AP-1 activation.
EMBO Rep 14:975-983 (2013). Garvin AJ, RM Densham, SA Blair-Reid, KM Pratt, HR Stone, D Weekes, . . . JR Morris. The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair.