Bioinformatics & Health Data Science

We are a multidisciplinary group with diverse expertise, backgrounds and skills ranging from biology, genetics, medicine, health informatics, clinical bioinformatics, genetic epidemiology, public health, epidemiology, computational biology, to data science, computer science, engineering, Artificial Intelligence, machine learning, big data, statistics and mathematics collaborating to address important questions in medical sciences.
Theme Centres
The Centre for Health Data Science, led by Professor Georgios Gkoutos and Professor Krishnarajah Nirantharakumar includes clinicians and academics that cover expertise in digital epidemiology, clinical bioinformatics, learning health systems, and artificial intelligence for health. With research and training at the core of our Centre, we aim to harness the full potential of Birmingham’s health data science capabilities to develop regional, national, and global leadership in the field of health data science.
The Centre for Environmental Research and Justice (CERJ) (Deputy Director: Professor Georgios Gkoutos) brings together academic expertise across three university colleges – Life and Environmental Sciences, Arts and Law, Medical and Dental Sciences – with an ambitious mission to help remedy harm to human health and the environment caused by pollution.
Theme Lead

Professor Georgios Gkoutos
Director - Centre for Health Science
Deputy Director - Centre for Environmental Research and Justice
Chair of Clinical Bioinformatics
View Professor Gkoutos' profile
Research groups
| PI | Research Interests |
|---|
|
Dr Animesh Acharjee, Assistant Professor of Integrative Analytics and AI
|
Multi-modal Integration, Diagnostics, Network Biology, Microbiomics |
| Dr Roland Arnold, Group Leader |
Bioinformatics analyses of transcription in cancer |
| Dr Deena Gendoo, Associate Professor of Computational Biology |
Bioinformatics characterization of preclinical disease models and their applications in therapy |
| Professor Georgios V. Gkoutos, Chair of Clinical Bioinformatics |
Clinical Bioinformatics, Health Data Science, Precision Medicine, Translational Phenomics |
| Dr Marc Haber, Associate Professor of Population Genetics, University of Birmingham Dubai |
Population Genetics |
| Dr Mohamed El-Hadidi, Associate Professor of Bioinformatics, University of Birmingham Dubai |
Bioinformatics |
| Dr Andreas Karwath, Associate Professor of Health Data Science |
Artificial intelligence and machine learning for health |
| Dr Anas Rana, Lecturer |
Statistical method development, ML methods |
| Dr Mamunur Rashid, Assistant Professor of Bioinformatics |
Bioinformatics |
| Dr Karin Slater, Assistant Professor of Biomedical Semantics |
Ontology, Semantics, Text mining, NLP, Artifical Intelligence |
| Dr Csilla Várnai |
Structural Bioinformatics |
Teaching and Education
The members of our section are responsible for the following programmes
In addition, we deliver a number of standalone modules for various programmes:
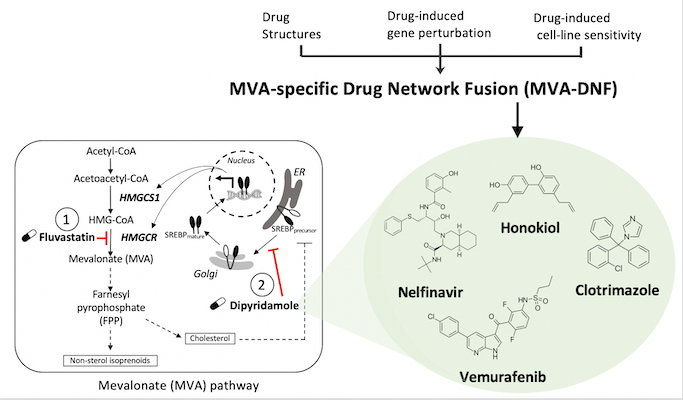
Spotlight on research using drug structure analysis to identify new therapeutics for breast cancer
In a study led by Deena Gendoo’s lab, an integrative pharmacogenomics pipeline was developed to identify novel and effective therapeutics for breast cancer therapy. This pipeline, mevalonate drug-network fusion (MVA-DNF), integrated drug-induced gene perturbation signatures, drug sensitivity, and drug structure profiles to generate a pathway-centric network of drugs that can target the mevalonate (MVA) pathway.
The mevalonate pathway is a metabolic pathway that is a hallmark of many cancer types, including breast cancer, as it produces cholesterol and other isoprenoids that are essential for cell proliferation and survival. Dipyridamole (DP) is a drug which synergizes with statins to potentiate the pro-apopotic activity of statins as part of the mevalonate pathway, but this drug has limited clinical utility for cancer patients. Using MVA-DNF, new statin-drug combinations were identified that target the MVA pathway, and these hits have been validated by testing on cancer cell-lines and patient-derived organoids, showing their potential for breast cancer therapy.
Accordingly, this work identified several actionable compounds and novel and effective therapeutics to help combat difficult-to-treat triple-negative breast cancer.
van Leeuwen J.E., Ba-Alawi W., Branchard E, Cruickshank J, Schormann W, Longo J, Silvester J, Gross P, Andrews D, Cescon D, Haibe-Kains B, Linda P, Gendoo D. Computational pharmacogenomic screen identifies drugs that potentiate the anti-breast cancer activity of statins. Nat Commun 13, 6323 (2022).
Spotlight on COVID-19 and Leukaemia
New study describes which cancer patients are more vulnerable to COVID-19.
Lancet Oncology 21: 1309-1316 (2020). Lee LYW, JB Cazier, T Starkey, SEW Briggs, R Arnold, V Bisht, . . . UKCCMP Team. COVID-19 prevalence and mortality in patients with cancer and the effect of primary tumour subtype and patient demographics: a prospective cohort study.
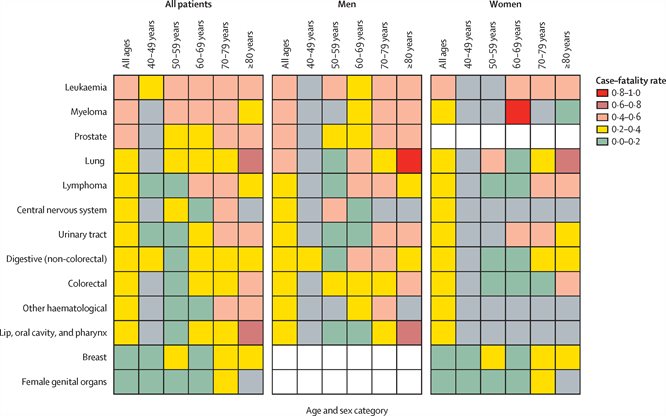
A newly published study involving Lennard Lee and Jean-Baptiste Cazier has found that, compared to other cancers, patients with blood cancers are more vulnerable to the effects of the coronavirus pandemic. The researchers were able to determine that patients with haematological cancers, particularly older patients and those with leukaemia, had a more severe COVID-19 trajectory compared to patients with solid organ tumours.
Selected Highlights from this Research Theme
Eur Heart J (2023). Gill SK, A Karwath, HW Uh, VR Cardoso, Z Gu, A Barsky, . . . GV Gkoutos, D Kotecha, BigData@Heart Consortium and the cardAIc group. Artificial intelligence to enhance clinical value across the spectrum of cardiovascular healthcare. doi: 10.1093/eurheartj/ehac758
Gut (2023). Karamitopoulou E, Wenning AS, Acharjee A, et al. Spatially restricted tumour-associated and host-associated immune drivers correlate with the recurrence sites of pancreatic cancer. doi:10.1136/gutjnl-2022-329371
JAMA Netw Open 5:e220130 (2022). Varnai C, C Palles, R Arnold, HM Curley, K Purshouse, VWT Cheng, . . . J-B Cazier, The UKCCMP Team. Mortality Among Adults With Cancer Undergoing Chemotherapy or Immunotherapy and Infected With COVID-19.
Cell Rep Med. (2022).Irvine HJ, Acharjee A, Wolcott Z, et al. Hypoxanthine is a pharmacodynamic marker of ischemic brain edema modified by glibenclamide. doi:10.1016/j.xcrm.2022.100654
Bioinformatics. (2022).Xu Y, Nash K, Acharjee A, Gkoutos GV. CACONET: a novel classification framework for microbial correlation networks. doi:10.1093/bioinformatics/btab879
JAMA Psychiatry (2022), Williams JA, Burgess S, Suckling J, Lalousis PA, Batool F, Griffiths SL, Palmer E, Karwath A, Barsky A, Gkoutos GV, Wood S, Barnes NM, David AS, Donohoe G, Neill JC, Deakin B, Khandaker GM, Upthegrove R; PIMS Collaboration. Inflammation and Brain Structure in Schizophrenia and Other Neuropsychiatric Disorders: A Mendelian Randomization Study., May 1;79(5):498-507. doi: 10.1001/jamapsychiatry.2022.0407.
Lancet 398:1427-1435 (2021). Karwath A, KV Bunting, SK Gill, O Tica, S Pendleton, F Aziz, . . .GV Gkoutos, D Kotecha, The cadrdAlc group and the Beta-blockers in Heart Failure Collaborative Group. Redefining beta-blocker response in heart failure patients with sinus rhythm and atrial fibrillation: a machine learning cluster analysis.
Cell 184:4612-4625 e4614 (2021). Almarri MA, M Haber, RA Lootah, P Hallast, S Al Turki, HC Martin, . . . C Tyler-Smith. The genomic history of the Middle East.
American J Human Genetics 107:149-157 (2020). Haber M, J Nassar, MA Almarri, T Saupe, L Saag, SJ Griffith, . . . C Tyler-Smith. A Genetic History of the Near East from an aDNA Time Course Sampling Eight Points in the Past 4,000 Years.
EClinicalMedicine (2021), Malik NS, Chernbumroong S, Xu Y, Vassallo J, Lee J, Moran CG, Newton T, Arul GS, Lord JM, Belli A, Keene D, Foster M, Hodgetts T, Bowley DM, Gkoutos GV., Paediatric major incident triage: UK military tool offers best performance in predicting the need for time-critical major surgical and resuscitative intervention. Aug 23;40:101100. doi: 10.1016/j.eclinm.2021.101100. eCollection 2021 Oct.
Nat Commun 11:1825 (2020). Chung PED, DMA Gendoo, R Ghanbari-Azarnier, JC Liu, Z Jiang, J Tsui, . . . E Zacksenhaus. Modeling germline mutations in pineoblastoma uncovers lysosome disruption-based therapy.
Am J Hum Genet 107:149-157 (2020). Haber M, J Nassar, MA Almarri, T Saupe, L Saag, SJ Griffith, . . . C Tyler-Smith. A Genetic History of the Near East from an aDNA Time Course Sampling Eight Points in the Past 4,000 Years.
Lancet Oncol 21:1309-1316 (2020). Lee LYW, JB Cazier, T Starkey, SEW Briggs, R Arnold, V Bisht, . . . UKCCMP Team. COVID-19 prevalence and mortality in patients with cancer and the effect of primary tumour subtype and patient demographics: a prospective cohort study.
EClinicalMedicine (2021), Malik NS, Chernbumroong S, Xu Y, Vassallo J, Lee J, Bowley DM, Hodgetts T, Moran CG, Lord JM, Belli A, Keene D, Foster M, Gkoutos GV. The BCD Triage Sieve outperforms all existing major incident triage tools: Comparative analysis using the UK national trauma registry population, May 15;36:100888. doi: 10.1016/j.eclinm.2021.100888.
Comput Biol Med. (2021).Bisht V*, Acharjee A*, Gkoutos GV. NFnetFu: A novel workflow for microbiome data fusion. Comput Biol Med. 2021;135:104556. doi:10.1016/j.compbiomed.2021.104556
Front Genet 11:669 (2020). Mountford HS, P Villanueva, MA Fernandez, L Jara, Z De Barbieri, LG Carvajal-Carmona, . . . JB Cazier, DF Newbury. The Genetic Population Structure of Robinson Crusoe Island, Chile.
Sci Rep 9:17405 (2019). Althubaiti S, A Karwath, A Dallol, A Noor, SS Alkhayyat, R Alwassia, . . . GV Gkoutos, R Hoehndorf. Ontology-based prediction of cancer driver genes.
Sci Data. (2019). Bravo-Merodio L, Acharjee A, Hazeldine J, …, Gkoutos GV, Lord J Machine learning for the detection of early immunological markers as predictors of multi-organ dysfunction. doi:10.1038/s41597-019-0337-6
PLoS Comput Biol 15:e1006596 (2019). Gendoo DMA, RE Denroche, A Zhang, N Radulovich, GH Jang, M Lemire, . . . B Haibe-Kains. Whole genomes define concordance of matched primary, xenograft, and organoid models of pancreas cancer.
J Transl Med (2019). Bravo-Merodio L, Williams JA, Gkoutos GV, Acharjee A. Omics biomarker identification pipeline for translational medicine. doi:10.1186/s12967-019-1912-5